NHGRI funding for HGNC and VGNC
We are delighted to confirm the renewal of our funding from the National Human Genome Research Institute (NHGRI) of the USA’s National Institutes of Health (U24HG003345). Together with our grant from the Wellcome Trust this will enable us to continue our human gene naming and also support the continuation of our VGNC work on naming genes across key vertebrate species that lack a dedicated nomenclature committee. Our NHGRI funding also provides subcontracts for our specialist advisors for the olfactory receptor and cytochrome P450 families and will allow us to investigate cross-vertebrate naming in other complex families such as the UDP glucuronyltransferases and soluble glutathione-S-transferases.
Please try our beta release at beta.genenames.org
Regular visitors to genenames.org will hopefully have noticed our requests to visit our new beta site at https://beta.genenames.org/. We would very much appreciate as many of our newsletter readers as possible to visit the beta site and to leave us feedback via our online form at https://www.genenames.org/cgi-bin/feedback. We would like all opinions and are keen to hear what you like as well as what you think could be improved!
The main reasons behind the website redesign are to make the site fully useable on mobile devices, improve our search application and to give the site a more modern appearance. We would be interested to hear how your experiences differ in looking at the beta site on mobile devices vs laptops and desktops.
Here is a summary of some of the main improvements:
The homepage
This has been redesigned to focus attention on the search box and to provide a simple description of the resource to inform those who visit the site for the first time, the layout being loosely based on the Google search homepage.
Gene symbol reports
New features in this redesign include:
- compatibility with mobile devices
- a data summary at the top in an orange box which contains the approved gene symbol, gene name, locus type and HGNC ID
- a separate tab called ‘HCOP homology predictions’ that goes to the HCOP homology search results for that particular gene
- easier viewing and pasting of information contained within a single field, such as within alias symbols
- the ability to open and close the main sections of the report, such as Gene Resources and Protein Resources
- a new ‘Orthologs from selected species’ section that includes, where applicable, the named VGNC orthologs for Bos taurus (cow), Equus caballus (horse), Pan troglodytes (chimp) and Canis familiaris (dog). This section also displays, where applicable, the mouse and rat orthologs with links out to the MGD and RGD databases.
- the ability to show/hide data using a +/- for some fields with multiple entries such as CCDS IDs, UniProt/SwissProt accessions and LSDB entries
- inclusion of new links to the AmiGO resource
- inclusion of new links to the Genetics Home Reference resource
Gene group reports
One major change is that we have stopped using the term ‘gene families’ and we have replaced this with the term ‘gene groups’. The change in name is to represent the varied nature of our gene groups - many of these such as ‘MTOR complex 1’, ‘Blood group antigens’ and ‘Ion channels’ are outwith the ‘normal’ definition of a gene family.
Improved features of the gene group reports include:
- the ability to open and close sections
- the ability to click on gene group boxes within the gene hierarchy map and be taken straight to the gene group of interest as a default
- a ‘rearrange mode’ radio button within the gene hierarchy map which allows moving of the gene group boxes around within the map - this is useful for rearranging complicated maps to take screenshots
- inclusion of HGNC IDs in the gene group tables
The Search
Our search function has been improved so that searching with an alias symbol will return relevant approved symbols higher up the search results page. For an example of this, search for p53 on our current site and then perform the same search on our beta site. The first result on the beta site is TP53 while TP53 is on page 2 of the search results on the current site. Unlike the current site, searching the beta site with gene symbols will not return hits to gene groups. We made this decision to increase the relevance of search results. To easily find which gene groups a gene belongs to you can follow the search results link to the gene report to view group membership there. Searching with gene group names and aliases will still return gene groups as results.
We have also improved the way search results are presented. We now use colour coding for the three main types of search result: Gene [orange], Group [dark blue], Other [grey]. These colours are used both in the search results list and in the search filter on the left hand side. When genes are returned in the search, the ‘Filter by locus group’ option is open by default underneath ‘Filter by type’. Please see the screenshot below showing the second page of results for the search term ‘kinase’:
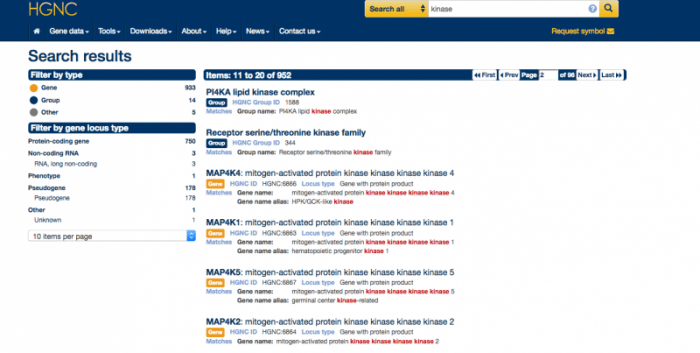
Please contact us with your feedback before the end of the summer. We plan to release the new website fully in the autumn.
New Gene Family Resources
The following gene families were added to our resource in the last couple of months:
Calmodulin dependent protein kinases
Chromatin accessibility complex
Granule associated serine proteases of immune defence
Spotlight on a new gene family:
We were recently contacted by a gene annotator asking us to name three novel protein coding genes present in a cluster with the genes OOSP1, OOSP2 and PLAC2. We set to work to determine the correct nomenclature for these genes and constructed our own phylogenetic tree to help us. As a result we named the genes OOSP3, OOSP4A and OOSP4B and we also added the symbol aliases OOSP2B to PLAC2 and OOSP2A to OOSP2 to reflect their paralogous relationship. You can view all of these genes on our new OOSP family page.
Gene Symbols in the News
We bring news of a possible explanation as to why some shift workers experience higher levels of exhaustion than others - researchers at the University of Helsinki found a common variant of MTNR1A is more often found in the exhausted shift workers compared to those who are less affected by their work patterns.
In other news, a study showed how the CRISPR technique can be used to assess whether a particular candidate gene variant has a pathological effect. In this case, the researchers found an individual who carried one copy of a particular variant of the MYL3 gene that had previously been implicated in increased risk of hypertrophic cardiomyopathy. The isolated individual had no symptoms, nor did any of his family members who carried the same variant. The researchers then edited induced pluripotent stem cells from this individual using the CRISPR technique to make either non-variant or stem cells homozygous for the MYL3 candidate variant. Studies on these edited cells suggested that this variant is not pathogenic.
Meeting News
This autumn Bryony and Elspeth are attending the Genome Informatics meeting in Hinxton from 17th-20th September, while Tamsin and Beth are attending the Livestock Genomics Meeting also in Hinxton on the 21st and 22nd September. Ruth will be travelling to Heidelberg in Germany to attend The Complex Life of RNA from 3rd-6th October. We will report back on our experiences of these travels in the next newsletter!
