Gene groups help
Introduction
We strongly encourage naming families and groups of genes related by sequence and/or function using a “root” symbol. This is an efficient and informative way to name related genes, and already works well for a number of established gene groups. Our gene groups data are now fully searchable via our search tool, please see search help for further information.
Gene groups index
Our gene groups index is ordered alphabetically according to group name, with root symbols shown in a separate column. Results are paginated and the user can choose to alter the number of results per page. Typing in either search box narrows down the results based on the input, as shown in the example below.
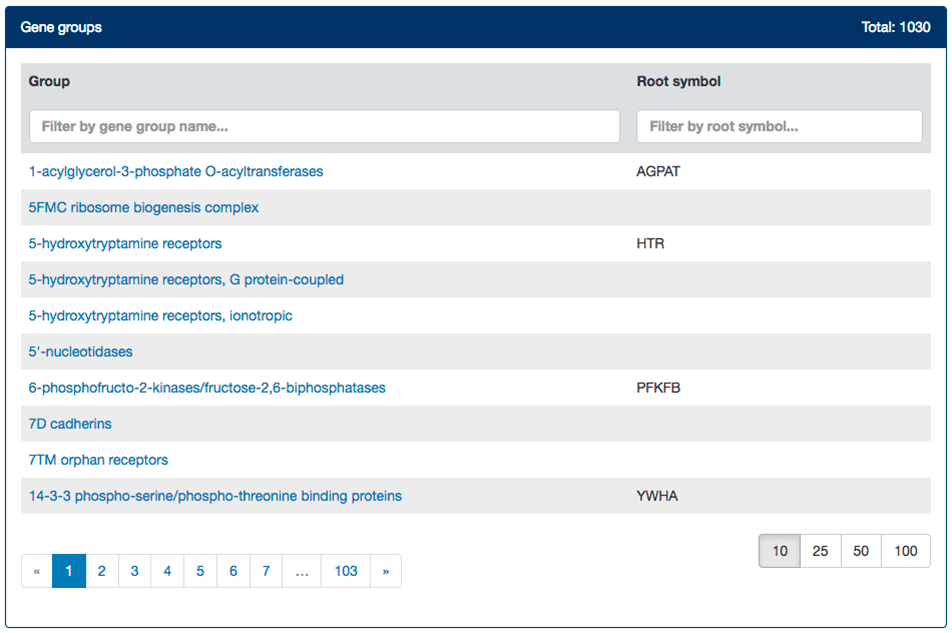
Gene groups pages
Our gene groups pages have been redesigned to provide more information, conform to a more standardised format, and make it easier to browse between groups. An example gene group page for Cholinergic receptors, muscarinic is shown below.
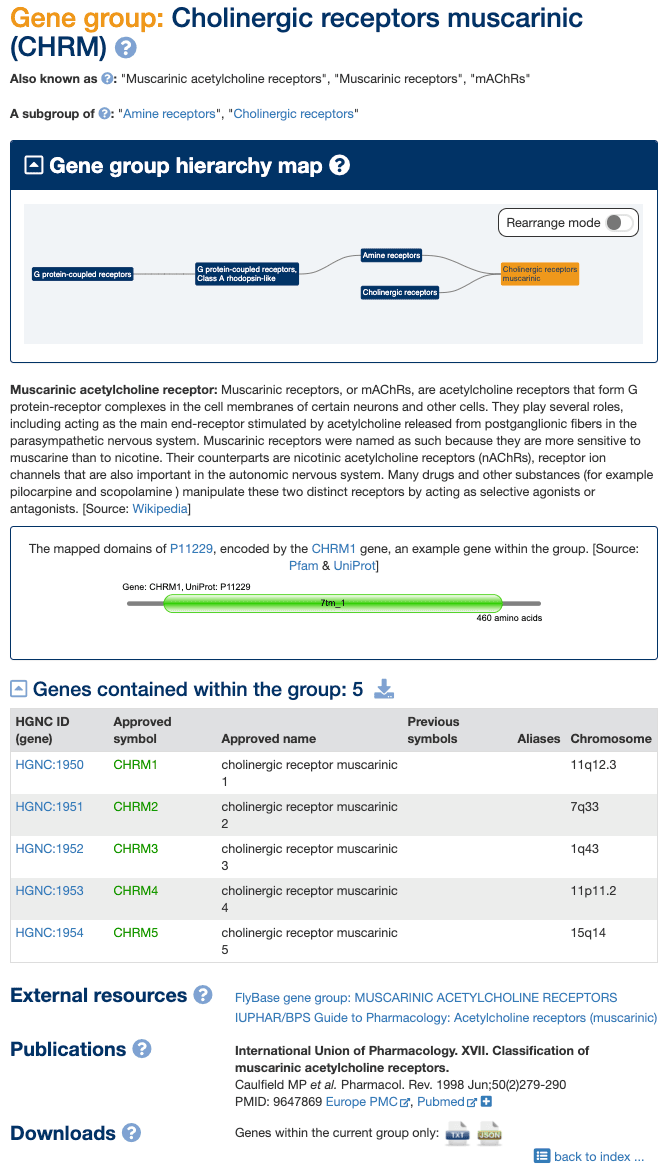
Gene group names, IDs and aliases
Each gene group has a unique numerical ID that forms the last part of the gene group page URL to aid linking and downloading. For example the numerical ID for “Cholinergic receptors, muscarinic” is 180. Each group also has a unique gene group name and, where a group has a root symbol, this is shown in parentheses next to the name. Please note that not all gene group pages equate to a particular set of genes with the same root symbol; in these cases only a gene group name is displayed. Other commonly-used gene group names and abbreviations are listed following the text “Also known as:” e.g. “Cholinergic receptors, muscarinic” are also known as “Muscarinic acetylcholine receptors”, “Muscarinic receptors” and “mAChRs”.
Gene group hierarchy map
The gene groups pages provide a display of curated hierarchical relationships between groups and allow users to browse easily through each hierarchy. All gene groups that fall into hierarchies include a "Gene group hierarchy map". By default each gene group box contains a link to that particular gene group page. The rearrange mode gives a user the option to move the boxes within the graphic, e.g. for use in a screenshot. Please note that the hyperlinks will no longer work when using this function. Clicking and holding the mouse button highlights the current gene group and its direct relatives, and highlights the path between them. To return to the default mode and to re-activate the links click on the rearrange mode switch once more.
In addition to the map, text links to any related groups are provided within the page. For example, in Fig 2 there are direct links to the Amine receptors and Cholinergic receptors.
Gene group descriptions
Many of our gene group pages contain a description of the group. These are often from Wikipedia (as shown in Fig 2) or UniProt (e.g. Integrins), in which case the source is clearly marked with a link through to the original page. In some cases the descriptions have been written by HGNC curators and these can be identified by [Source: HGNC] (e.g. IGH orphons); if they come from another source this will be clearly displayed within square brackets.
Example gene mapped domains graphic
Where gene group members share a particular protein domain we often show a graphical display of the protein domain structure for an example gene group member, which is sourced from Pfam via UniProt ID. In Fig. 2 the domain structure is shown for the product of the CHRM1 gene. Hovering over a domain within the graphic will reveal a label containing the domain name, description and Pfam family ID, while clicking on a domain takes the user through to the Pfam description page for that domain.
Genes within the group
HGNC Symbol Reports for each gene within a group can be accessed by clicking on the Approved Symbol. By default the table of group members is sorted by Approved Symbol, but where the group shares a root symbol the members can be sorted by that symbol even where it is a alias or previous symbol, e.g. in the DEAD box polypeptides (DDX) group INTS6 is sorted by its previous symbol of DDX26. Note that the symbol used for sorting is highlighted in green to make this clear.
Above the table of group members there is a small pie chart symbol next to the text “Genes contained within the group”. Clicking on the pie chart opens a pop-up box containing statistics on the locus types of the genes included in the group. For example, the Tubulin gene group contains 22 protein-coding genes and four pseudogenes.
Gene group downloads
We now provide a way of downloading gene groups as data sets, allowing users to choose between downloading a single group or the entire group hierarchy. For example, users can choose between downloading only the Cholinergic receptors, muscarinic genes shown in Fig 2 or they can browse through to the G protein-coupled receptors and download all genes belonging to that hierarchy. Each gene group page has a download link at the bottom of the page that generates a text file with all gene symbols and extra data fields such as “Approved Name” and “HGNC ID”. Please note that some gene group pages do not contain a list of genes because these are included to complete the hierarchical structure; these pages enable users to download all the genes from further down the hierarchy, e.g. Amine receptors and Cholinergic receptors.
Summary of core data fields
- Gene group name and root symbol
- At the top of the gene group report page we display the group name and if applicable the common root symbol of the genes associated with the gene group within rounded brackets.
- Also known as
- Synonymous names for the gene group.
- A subgroup of
- This field contains links to groups that the current group belongs.
- Group contains the following subgroups
- Contains links to sub groups within the current group.
- Comments
- This field contains useful information about the gene group supplied by one of our curators.
- Specialist advisors
- Names of specialists that advise and recommend appropriate gene symbols to the committee for a particular gene group.
- External resources
- Links to resources that will provide extra information about the current group.
- Publications
- References pertinent to the gene group. The user can choose to view these references at either PubMed or European PubMed Central. This section does not aim to list all possible published papers on the group but provides links to papers that first described the gene group in question or papers that are particularly relevant to the nomenclature of the genes.
- Downloads
- Download gene group data in a csv text format.