Search help
Please note: You no longer have to use the wildcard * when searching via a symbol. This makes it much easier to search our database of gene symbol reports using a root/prefix of a symbol such as ZNF (for Zinc finger gene) or OR (for Olfactory receptors). Our search will also give suggestions while you type and by clicking on a suggestion will take you straight to the record rather than having to use the search results interface.
Our search allows users to search all of our active gene symbol reports, gene group reports and static web pages quickly and with ease. The search server utilises Apache Solr which offers a powerful full-text search, hit highlighting and faceted searching. Faceted searching allows you to search for a particular keyword and then filter the results by record/page type, locus group and locus type.
The search form can be found within the mast head of each page. Enter a search term within the search input box and click on the spy glass icon. When typing into the inbox field, suggestions may appear. Selecting a suggestion will take you to the record bypassing the search result from for a speedier service. Please remember that this is a suggestion rather than the full result of a search. To retrieve the full result please don't select a suggestion and press either entery or click on the spy glass icon.
Basic search
The simplest way to search is to type a query word/ID into the input box within the mast head and click on the spy glass. The default option is a full-text search over all the indexes and fields. If reports are found containing the keyword/ID they are displayed in order of relevance (for more information see indexed fields for each search type). On the left hand side of the results page the filter options (where available) are shown with the numbers of reports associated with each facet. If the results include gene symbols then clicking on the facet "Gene" will filter the results by this type and will change the faceting to display the locus groups and types that are relevant to the search results; enabling further filtering by locus type. Users can also change the default number of results per page from 10 up to 200 and download the results from the search in either TXT (tab separated) or JSON format.
The results display specific fields from within the search index which differ depending on the document type. The first line of each result contains the gene symbol and the gene name if the result is a gene symbol report, the group name if it's a gene group report or the page title if it hits any other page within the site. The second row will show the type of the indexed document (i.e gene, group or site) if searching everything, and will also contain some of the important fields to help identify the hit. The third row reports the field the keyword/ID matches, so if the keyword matched an approved symbol within a gene symbol report the third row would say "Matches: Gene symbol etc" as seen below in figure 1.
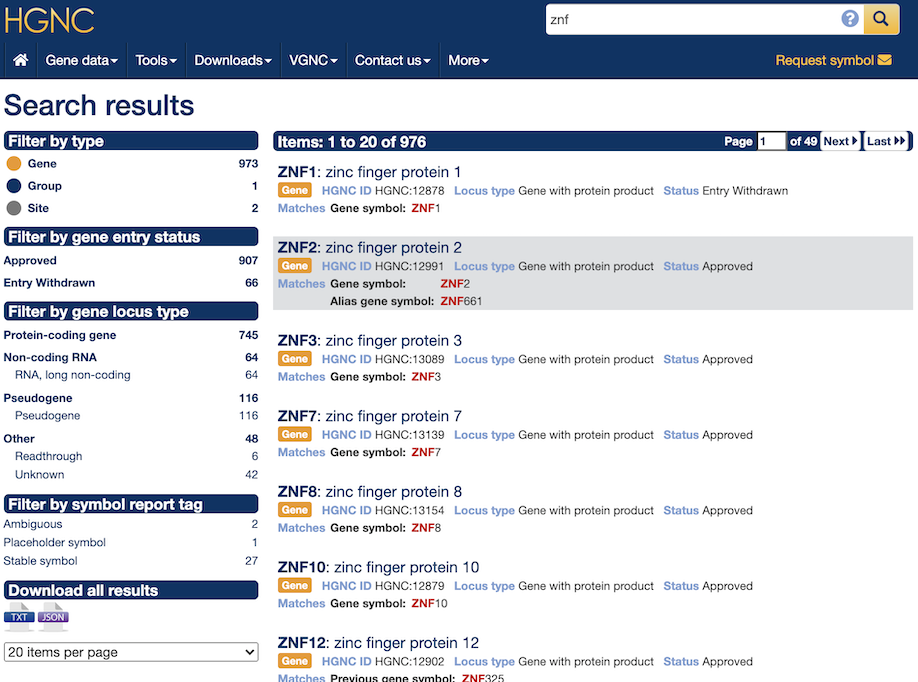
Advanced search
The search application allows users to make advanced queries using the search box. In this section we describe how to specify the search type, use wildcards, logic operators and specify indexed fields.
Search types
Instead of searching everything, users can select to search only for gene symbol reports, gene groups or the static content of the site (e.g. help or news pages) by selecting these options from the dropdown next to the search input box within the mast head.
Wildcard search
Sometimes it may be useful to match records based on a query pattern rather than a keyword or ID. Our search allows users to use wildcard operators with an asterisk (*) to stand in place for one or more characters and a question mark (?) to stand in place for a single character substitution. Multple wildcards can be used in the same query, for instance searching for TP?T* will find the symbols TPMT, TPST1, TPST2P1.
Logic operators
By default the search application uses the logical operator OR, so inputing BRAF RNA into the search box actually equates to BRAF OR RNA so some of the results will contain BRAF and some will contain RNA and others may contain both. Sometime however this isn't what a user wants to search. Lets say a user wants to find reports that contain both BRAF and RNA in the same report. Typing in BRAF RNA will unfortunately return over 7000 hits. By changing the search query to BRAF AND RNA the results returned are more pertinent and reduces the number of hits returned. Alternatively asking for reports that do not contain a keyword/ID may be preferable therefore users may use NOT or - within term such as BRAF NOT RNA or BRAF -RNA.
Phrases
As discussed above the default operator is OR, so if a search using the term protein arginine methyltransferase 1 was used the actual search term will be protein OR arginine OR methyltransferase OR 1. This can be addressed by simply quoting the query so that the search knows to treat the quoted block as one term like "protein arginine methyltransferase 1".
Indexed fields
Users may search within a specific indexed field by using the information seen in the indexed fields section below using a very basic notation. To specify an index the user need only to type in to the search field the indexed field key followed immediately with a colon (:) and then the query eg refseq_accession:NM_033360. If the query is not an ID or a keyword "phrases" can be used after the colon eg alias_gene_name:"A-kinase anchor protein, 350kDa".
Indexed fields
- agr
- Alliance of Genome Resources HGNC ID. This field may be found within the "Gene resources" section of the gene symbol report e.g. HGNC:11998
- alias_gene_name
- Other names used to refer to a gene as seen in the "Alias names" field in the gene symbol report e.g. A-kinase anchor protein, 350kDa
- alias_gene_symbol
- Other symbols used to refer to a gene as seen in the "Alias symbols" field in the gene symbol report e.g. BRAF1
- alias_group_name
- Other names used to refer to a group as seen within the gene group reports under the "Also known as" field e.g. LPAAT
- ccds_id
- Consensus CDS ID. This field may be found within the "Nucleotide resources" section of the gene symbol report e.g. CCDS5863
- curator_note
- Contains additional information related to a gene symbol report that has been manually added by an HGNC curator
- ena
- International Nucleotide Sequence Database Collaboration (GenBank, ENA and DDBJ) accession number(s). Found within the "Nucleotide resources" section of the gene symbol report e.g. M95712
- ensembl_gene_id
- Ensembl gene ID. Found within the "Gene resources" section of the gene symbol report e.g. ENSG00000157764
- gene_name
- HGNC approved name for the gene. Equates to the "Approved name" field within the gene symbol report e.g. zinc finger protein 536
- gene_symbol
- The HGNC approved gene symbol. Equates to the "Approved symbol" field within the gene symbol report e.g. KLF4
- group_id
- The HGNC gene group ID which can be seen in the URL for a gene group page after the word "group" e.g. 46
- group_name
- The gene group name as set by the HGNC and seen at the top of the gene group reports e.g. 1-acylglycerol-3-phosphate O-acyltransferases
- hgnc_id
- HGNC ID. A unique ID created by the HGNC for every approved symbol e.g. HGNC:1097
- locus_group
- A group name for a set of related locus types as defined by the HGNC e.g. non-coding RNA.
- locus_type
- The locus type as set by the HGNC e.g.RNA, long non-coding
- mane_select
- MANE Select nucleotide accession with version (i.e. NCBI RefSeq or Ensembl transcript ID and version). Found within the "Nucleotide resources" section of the gene symbol report e.g. NM_001374258.1 or ENST00000644969.2
- mgd_id
- Mouse genome informatics database ID. Found within the "Homologs" section of the gene symbol report e.g. MGI:88190
- ncbi_gene_id
- NCBI gene ID. Found within the "Gene resources" section of the gene symbol report e.g. 673
- omim_id
- OMIM ID. Found within the "Clinical resources" section of the gene symbol report e.g. 164757
- page_content
- Text contained within the static web page (i.e text within news or help pages) e.g. "human TBX18 gene"
- page_title
- The title of the static web page (i.e text within news or help pages) e.g. "TBX18 gene therapy"
- page_url
- The URL of the indexed static web page (i.e text within news or help pages) e.g. hgnc-newsletter-winter-2012-2013
- prev_gene_name
- Gene names previously approved by the HGNC for a gene. Equates to the "Previous names" field within the gene symbol report e.g. solute carrier family 5 (choline transporter), member 7
- prev_gene_symbol
- Gene symbols previously approved by the HGNC for this gene. Equates to the "Previous symbols" field within the gene symbol report e.g. RN5S49
- rgd_id
- Rat genome database gene ID. Found within the "Homologs" section of the gene symbol report e.g. RGD:2981
- refseq_accession
- RefSeq nucleotide accession. This field may be found within the "Nucleotide resources" section of the gene symbol report e.g. NM_033360
- root_symbol
- The common root gene symbol associated to a group if a common root symbol exists.
- status
- Status of the symbol report, which can be either "Approved" or "Entry Withdrawn" e.g. Approved
- symbol_report_tag
- The symbol report tags are tags/labels that may appear next to the symbol at the top of a symbol report. The value must be either "Ambiguous", "Placeholder symbol" or "Stable symbol" in the case shown.
- ucsc_id
- UCSC gene ID. Found within the "Gene resources" section of the gene symbol report e.g. uc002qsd.5
- uniprot_id
- UniProt protein accession. Found within the "Protein resource" section of the gene symbol report e.g. P00568