HCOP help
The HGNC Comparison of Orthology Predictions (HCOP) search is a tool that integrates and displays the orthology assertions predicted for a specified human gene, or set of human genes, by eggNOG, Ensembl Compara, HGNC, HomoloGene, Inparanoid, NCBI Gene Orthology, OMA, OrthoDB, OrthoMCL, Panther, PhylomeDB, PomBase, TreeFam and ZFIN. An indication of the reliability of a prediction is provided by the number of databases which concur. HCOP was originally designed to show orthology predictions between human and mouse, but has been expanded to include data from chimp, macaque, rat, dog*, horse, cattle, pig, opossum, platypus, chicken, anole lizard, xenopus, zebrafish, C. elegans, Drosophila, S. cerevisiae, and S. pombe meaning that there are currently 19 genomes available for comparison in HCOP.
*In release 105 of Ensembl the default dog assembly was changed from CanFam3.1 (a boxer) to ROS_Cfam_1.0 (a labrador retriever). As HCOP aims to be as comprehensive as possible it will include orthology data predicted using sequences from both dog genome assemblies until all the orthology sources have updated to use the Ros_Cfam_1.0 assembly data, at which point orthology predictions from CanFam3.1 will be retired. HCOP results use two different dog icons to allow users to easily distinguish which assembly the orthology prediction comes from.
Using the search
Orthology assertions can be obtained for a gene by searching with its Ensembl gene identifier or its NCBI Gene identifier. For species with either a model organism or nomenclature database you can also search with an approved gene symbol, an approved gene name or the gene identifier from that database. The species of the query gene must be selected using the drop down menu in the “Search for ortholog(s) between;” section of the form, and one or more species for which you wish to see orthology data should be selected using the species check boxes just below this. If your query is not a human gene you will only be able to identify orthologs beween the species you have provided gene information for and human. In the latest version of HCOP you can now specify which orthology sources you wish to see represented in the results. To include or exclude particular sources use the checkboxes in the ‘include orthologs from’ section of the form. If you choose to exclude a certain source then orthologs assignments made based only on evidence from that source will be excluded from the result set. You will still see assignments made by the excluded source appearing in results where the orthology assignment is supported by one or more of the other ortholog sources you have selected to use for the search. Not all sources have data for every species see the section on ‘origins of the data’ to find out what data are available. The results provide basic data about the query and its predicted homologs as well as a list of databases that support the assertion and links to further information.
The consensus orthology assertions for multiple genes can be viewed simultaneously by searching with a list of query terms, separated by commas, newlines or spaces. This list may either be pasted into the ‘enter identifier(s)’ box or uploaded as a file using the ‘upload file’ option on the form. If you have entered information into the ‘enter identifiers’ text box but then decide you would rather upload a file containing your data you must either delete the data in the text box or clear the form using the ‘reset’ button before uploading your file of data.
If provided with a list of terms (either comma, space or newline delimited), HCOP searches with each term in turn e.g. ABCA1 ABCA2 ABCA3. A separate result panel will appear for each query term that produces a result. If more than one query term was supplied the results sections may be scrollable.
HCOP supports wild cards. Use _ to substitute for a single character and * or % to substitute for 1 or more characters. For example ABCA* fetches all genes beginning ABCA, while ABCA_ fetches only ABCA1 to ABCA9. It is not advisable to start a query with a wildcard as the database will not be able to use its indexes and the search will be slow.
Searches are case insensitive. The ‘HGNC:’, ‘VGNC:’, ‘MGI:’, ‘RGD:’, ‘ZFIN’, ‘SGD:’, ‘XENBASE’, ‘BGD:’ or ‘CGNC:’ database identifier prefixes are not required, but the search will work whether they are included or not.
Search results
The latest version of HCOP has changed the way in which the results are displayed to make it more user friendly. Each query term that has an ortholog assignment in one or more of the ortholog species selected will have a results panel like this:
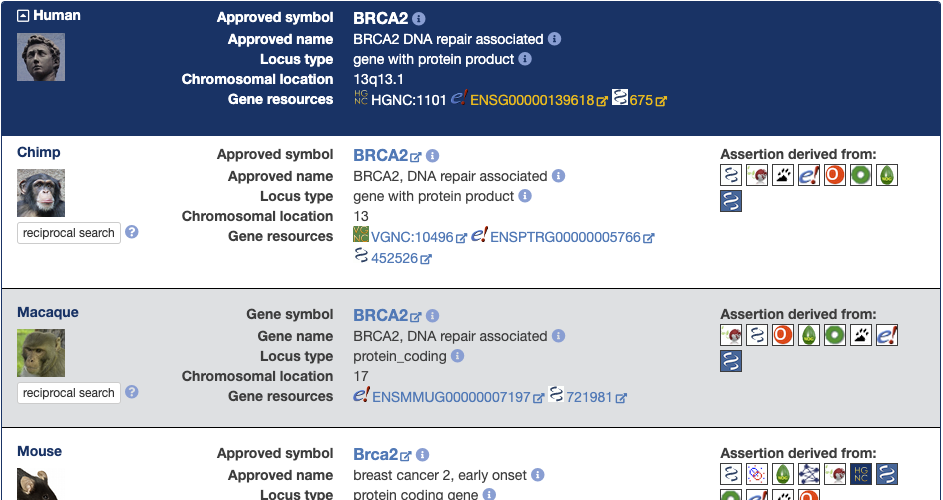
At the top of the results panel in the blue section you will see data relating to the query term you supplied. Below this you will see a section for each ortholog that has been returned. For the both the query and orthologs you should see:
Gene symbol
This will either be prefixed with ‘Approved symbol’ to indicate that the symbol was assigned by a nomenclature committee or with ‘Gene symbol’ to indicate that it is not an approved symbola non-approved symbol may in some cases be listed as ‘Unknown’. If you hover your mouse over the information icon to the left of the symbol a popup will tell you where we got the symbol data from.
Gene name
Again this will be prefixed with ‘Approved name’ to indicate that the name was assigned by a nomenclature committee or with ‘Gene name’ to indicate that this is not an approved name. If you hover your mouse over the information icon to the left of the name a popup will tell you where we got the name data from.
Locus type
This is the locus type of the gene; if you hover your mouse over the information icon to the left of the name a popup will tell you where we got the locus type information from.
Chromosomal location
Indicates the cytogenetic location of the gene or region on the chromsome. In the absence of that information one of the following may be listed:
- not on reference assembly - named gene is not annotated on the current version of the Genome Reference Consortium human reference assembly; may have been annotated on previous assembly versions or on a non-reference human assembly
- unplaced - named gene is annotated on an unplaced/unlocalized scaffold of the human reference assembly
- reserved - named gene has never been annotated on any human assembly
Gene resources
Links to the gene in its model organism or nomenclature database, Ensembl and NCBI Gene where applicable.
For each ortholog gene you will also see a section labelled ‘Assertion derived from:’ that contains a series of icons. These icons represent the ortholog sources that support this orthology assignement. Clicking on the icon will take you to the entry for that gene in the orthology database, while hovering over the icon will give you the full name of the ortholog data source.
Origins of the data
The data behind HCOP is stored in a MySQL database to allow for rapid querying. Each orthology assignment is stored as a pair of genes, with mapped database identifiers and basic gene data as well as a list of associated databases that support that assertion. The data in HCOP is updated weekly by running a pipeline that first works out if we have new data from any of our orthology sources and then updates the data as required. We currently import gene data from:
- Ensembl
- HGNC
- MGI
- NCBI Gene
- PomBase
- RGD
- SGD
- VGNC
- WormBase
- Xenbase &
- ZFIN
These data are used to ensure that we have the current approved gene symbols, names, locus types and location information from the appropriate nomenclature or model organism database. For those species without a nomenclature committee (currently this applies to chimp, macaque, dog, horse, cattle, pig, opossum and platypus) or where a nomenclature resource exists but gene data are not available (anole lizard) in a form we can use in our pipeline we take this information from the NCBI Gene database or from Ensembl if the gene in question can not be mapped to an NCBI Gene identifier.
Orthology data are imported from various sources, the table below details the sources we use, the current version of the data and the species this applies to:
| Orthology Source | Version | Species data applies to |
|---|---|---|
| eggNOG | Version 5.0 | All species except horse |
| Ensembl | Release 109 | All species |
| HGNC | N/A | Human and mouse |
| HomoloGene | Release 68 | Human, chimp, macaque, mouse, rat, dog, cattle, chicken, xenopus, zebrafish, C. elegans, fruitfly, S. cerevisiae & S. pombe |
| Inparanoid | Version 8.0 | All species |
| OMA | Release November 2022 | All species |
| OrthoDB | Version 11 | Human, chimp, macaque, mouse, rat, dog, cat, horse, cattle, pig, opossum, platypus, chicken, anole lizard, xenopus, zebrafish, C. elegans & fruitfly |
| OrthoMCL | Version 6.15 | Human, mouse, dog, chicken, xenopus, zebrafish, C. elegans, fruitfly, S. cerevisiae & S. pombe |
| NCBI gene orthology | N/A | All species except C. elegans, fruitfly, S. cervisiae & S. pombe |
| Panther | Version 17.0 | All species |
| PhylomeDB | Version 4, data are taken from phylome 514 | Human, chimp, macaque, mouse, rat, dog, cattle, opossum, platypus, chicken, xenopus, zebrafish, C. elegans, fruitfly, S. cerevisiae & S. pombe |
| PomBase | N/A | Human & S. pombe |
| TreeFam | Release 9.0 | All species except cat & S. pombe |
| ZFIN | N/A | Human & zebrafish |
For versioned orthology sources the pipeline used for updating HCOP will detect that the version of the data has changed and update the data accordingly. For those ortholog sources where no data version is specified the data will be updated each time the HCOP pipeline is run (currently weekly), so that HCOP will never be more than one week out of sync with the data source.
If there is an orthology source that you would like to see in HCOP please contact us via our feedback form and we will see if it is possible for it to be incorporated.
Linking to a HCOP result
It is now possible to go directly to an HCOP result using a URL with set parameters; this enables you to add links to our HCOP results within your own web applications. Use the following URL template:
https://www.genenames.org/tools/hcop/#!/?q=<query>&qtype=<query type>&qtax_id=<query NCBI taxon ID>&ttax_id=<target species NCBI taxon ID>&submit=true
e.g. Probable chimp and macaque orthologs of a human gene with the approved symbol of BRCA2.
https://www.genenames.org/tools/hcop/#!/?q=BRCA2&qtype=symbol&qtax_id=9606&ttax_id=9598&ttax_id=9544&submit=trueor
Probable human orthologs of the chimp gene with the Ensembl gene ID of ENSPTRG00000005766.
https://www.genenames.org/tools/hcop/#!/?q=ENSPTRG00000005766&qtype=ensembl&qtax_id=9598&ttax_id=9606&submit=true
Below is a table that will help you to fill in the ‘query type’ and taxon IDs. If the qtax_id is not human (9606) then only one ttax_id can be used and must be 9606.
| Species | NCBI taxon ID | Allowed query types for species |
|---|---|---|
| Human | 9606 | symbol, name, ensembl (i.e Ensembl gene ID), egene (i.e NCBI gene ID), hgnc (i.e HGNC ID) |
| Chimp | 9598 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Macaque | 9544 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Mouse | 10090 | symbol, name, ensembl, egene, mgi (i.e MGI ID) |
| Rat | 10116 | symbol, name, ensembl, egene, rgd (i.e RGD ID) |
| Dog | 9615 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Cat | 9685 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Horse | 9796 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Cattle | 9913 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Pig | 9823 | symbol, name, ensembl, egene, vgnc (i.e VGNC ID) |
| Opossum | 13616 | ensembl, egene |
| Platypus | 9258 | ensembl, egene |
| Chicken | 9031 | symbol, name, ensembl, egene |
| Anole lizard | 28377 | ensembl, egene |
| Xenopus | 8364 | symbol, name, ensembl, egene, xenbase (i.e XENBASE ID) |
| Zebrafish | 7955 | symbol, name, ensembl, egene, zfin (i.e ZFIN ID) |
| C.elegans | 6239 | symbol, name, ensembl, egene, wormbase (i.e WormBase ID) |
| Fruitfly | 7227 | symbol, name, ensembl, egene, flybase (i.e FlyBase ID) |
| S.cerevisiae | 4932 | symbol, name, ensembl, egene, sgd (i.e SGD ID) |
| S.pombe | 284812 | ensembl, pombase (i.e PomBase ID) |
Using the Bulk downloads tool
For your convenience we have pre-calculated some files of HCOP data that you can download from our FTP site. You have the option of getting a file containing human and ortholog data from a single species, or human and ortholog data from all HCOP species in a single file. For the human - single ortholog species files the ‘6 Column’ output returns the raw assertions, Ensembl gene IDs and Entrez Gene IDs for human and one other species, while the ‘15 Column’ output includes additional information such as the chromosomal location, accession numbers and where possible references the approved gene nomenclature.
The files containing all species ortholog data have an additional column at the start giving the taxon id for each ortholog species.
Referencing the HGNC Comparison of Orthology Predictions search tool
If you use this tool or the ‘Bulk Downloads’ in published work please reference:
- Yates B, Gray KA, Jones TEM, Bruford EA. Updates to HCOP: the HGNC comparison of orthology predictions tool. Brief Bioinform. 2021 May 6. PMID: 33959747 DOI: 10.1093/bib/bbab155
- Wright MW, Eyre TA, Lush MJ, Povey S and Bruford EA. HCOP: The HGNC Comparison of Orthology Predictions Search Tool. Mamm Genome. 2005 Nov; 16(11):827-828. PMID:16284797 PDF
- Eyre TA, Wright MW, Lush MJ and Bruford EA. HCOP: a searchable database of human orthology predictions. Brief Bioinform. 2007 Jan;8(1):2-5. PMID:16951416
- Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011 Jan;39(Database issue):D519-9. PMID: 20929869 PDF
Image credits
Images used in HCOP are either used with permission from the image owner, or are in the public domain.
- Species images are taken from Ensembl, see here for more details.
- The MGI logo is used with their permission
- The RGD logo is used with their permission
- The ZFIN logo is used with their permission
- The Ensembl logo is used with their permission
- The Homologene and NCBI logos are used with permission of the NCBI
- The Inparanoid logo is used with their permission
- The OMA logo is used with their permission
- The OrthoDB logo is used with their permission
- The Panther logo is used with their permission
- The PomBase logo is used with their permission
- The Treefam logo is used with their permission
- The WormBase logo is used with their permission